Relational Data and Joins
Learning Outcomes
- Describe relational data.
- Use the correct R tidyverse function to manipulate data:
inner_join(),left_join(),right_join(),full_join(),semi_join(),anti_join().
Relational Data
- Load the tidyverse
library(tidyverse)
- Many datasets (especially those drawn from relational databases) have more than two data frames.
- These data frames are often logically related where rows in one data frame correspond to, or, have a relation to, rows in another data frame.
- Large Relational Databases are typically designed (by data engineers) to achieve some level of “normalization”
- The goal of normalization is to design the set of tables (data frames) to capture all of the data while achieving a balance across:
- storage size,
- query speed,
- ease of maintenance by users, and
- minimal delays for distributed operations with multiple users.
- Table design strives to:
- reduce data duplication (which increases storage and the potential for errors), and
- enable multiple users to update their tables without interference from other users.
- A principle of normalization is to limit tables to a single purpose.
- As R users, we might say, a table (dataframe) describes one type of (observational or experimental) unit, with all the observations for each variable associated with that unit and only that unit.
- In modern databases, you may run into tables containing blob objects such as pictures, figures, or movies
- You may need to work with the database administrator to get only the data you need into R.
Example: The Lahman Data Set has Multiple Tables
- Consider the tables/data frames in the lahman package.
install.packages("Lahman")
library(Lahman)
- Use the
data(package = "package-name")to see the data sets in a package
data(package = "Lahman")
Each Table has One Purpose: To Describe One Type of Unit
# Let's turn these 5 dataframes into tibbles while we're here:
People <- as_tibble(Lahman::People)
Teams <- as_tibble(Lahman::Teams)
Fielding <- as_tibble(Lahman::Fielding)
Pitching <- as_tibble(Lahman::Pitching)
Batting <- as_tibble(Lahman::Batting)
# `People`: Player names, DOB, and biographical info.
head(People)
## # A tibble: 6 x 26
## playerID birthYear birthMonth birthDay birthCountry birthState birthCity
## <chr> <int> <int> <int> <chr> <chr> <chr>
## 1 aardsda~ 1981 12 27 USA CO Denver
## 2 aaronha~ 1934 2 5 USA AL Mobile
## 3 aaronto~ 1939 8 5 USA AL Mobile
## 4 aasedo01 1954 9 8 USA CA Orange
## 5 abadan01 1972 8 25 USA FL Palm Bea~
## 6 abadfe01 1985 12 17 D.R. La Romana La Romana
## # ... with 19 more variables: deathYear <int>, deathMonth <int>,
## # deathDay <int>, deathCountry <chr>, deathState <chr>, deathCity <chr>,
## # nameFirst <chr>, nameLast <chr>, nameGiven <chr>, weight <int>,
## # height <int>, bats <fct>, throws <fct>, debut <chr>, finalGame <chr>,
## # retroID <chr>, bbrefID <chr>, deathDate <date>, birthDate <date>
# `Teams`: Yearly statistics and standings for teams.
head(Teams)
## # A tibble: 6 x 48
## yearID lgID teamID franchID divID Rank G Ghome W L DivWin WCWin
## <int> <fct> <fct> <fct> <chr> <int> <int> <int> <int> <int> <chr> <chr>
## 1 1871 NA BS1 BNA <NA> 3 31 NA 20 10 <NA> <NA>
## 2 1871 NA CH1 CNA <NA> 2 28 NA 19 9 <NA> <NA>
## 3 1871 NA CL1 CFC <NA> 8 29 NA 10 19 <NA> <NA>
## 4 1871 NA FW1 KEK <NA> 7 19 NA 7 12 <NA> <NA>
## 5 1871 NA NY2 NNA <NA> 5 33 NA 16 17 <NA> <NA>
## 6 1871 NA PH1 PNA <NA> 1 28 NA 21 7 <NA> <NA>
## # ... with 36 more variables: LgWin <chr>, WSWin <chr>, R <int>, AB <int>,
## # H <int>, X2B <int>, X3B <int>, HR <int>, BB <int>, SO <int>, SB <int>,
## # CS <int>, HBP <int>, SF <int>, RA <int>, ER <int>, ERA <dbl>, CG <int>,
## # SHO <int>, SV <int>, IPouts <int>, HA <int>, HRA <int>, BBA <int>,
## # SOA <int>, E <int>, DP <int>, FP <dbl>, name <chr>, park <chr>,
## # attendance <int>, BPF <int>, PPF <int>, teamIDBR <chr>,
## # teamIDlahman45 <chr>, teamIDretro <chr>
# `Fielding`: Fielding stats
head(Fielding)
## # A tibble: 6 x 18
## playerID yearID stint teamID lgID POS G GS InnOuts PO A E
## <chr> <int> <int> <fct> <fct> <chr> <int> <int> <int> <int> <int> <int>
## 1 abercda~ 1871 1 TRO NA SS 1 1 24 1 3 2
## 2 addybo01 1871 1 RC1 NA 2B 22 22 606 67 72 42
## 3 addybo01 1871 1 RC1 NA SS 3 3 96 8 14 7
## 4 allisar~ 1871 1 CL1 NA 2B 2 0 18 1 4 0
## 5 allisar~ 1871 1 CL1 NA OF 29 29 729 51 3 7
## 6 allisdo~ 1871 1 WS3 NA C 27 27 681 68 15 20
## # ... with 6 more variables: DP <int>, PB <int>, WP <int>, SB <int>, CS <int>,
## # ZR <int>
# `Pitching`: Pitching stats
head(Pitching)
## # A tibble: 6 x 30
## playerID yearID stint teamID lgID W L G GS CG SHO SV
## <chr> <int> <int> <fct> <fct> <int> <int> <int> <int> <int> <int> <int>
## 1 bechtge~ 1871 1 PH1 NA 1 2 3 3 2 0 0
## 2 brainas~ 1871 1 WS3 NA 12 15 30 30 30 0 0
## 3 fergubo~ 1871 1 NY2 NA 0 0 1 0 0 0 0
## 4 fishech~ 1871 1 RC1 NA 4 16 24 24 22 1 0
## 5 fleetfr~ 1871 1 NY2 NA 0 1 1 1 1 0 0
## 6 flowedi~ 1871 1 TRO NA 0 0 1 0 0 0 0
## # ... with 18 more variables: IPouts <int>, H <int>, ER <int>, HR <int>,
## # BB <int>, SO <int>, BAOpp <dbl>, ERA <dbl>, IBB <int>, WP <int>, HBP <int>,
## # BK <int>, BFP <int>, GF <int>, R <int>, SH <int>, SF <int>, GIDP <int>
# `Batting`: Batting stats
head(Batting)
## # A tibble: 6 x 22
## playerID yearID stint teamID lgID G AB R H X2B X3B HR
## <chr> <int> <int> <fct> <fct> <int> <int> <int> <int> <int> <int> <int>
## 1 abercda~ 1871 1 TRO NA 1 4 0 0 0 0 0
## 2 addybo01 1871 1 RC1 NA 25 118 30 32 6 0 0
## 3 allisar~ 1871 1 CL1 NA 29 137 28 40 4 5 0
## 4 allisdo~ 1871 1 WS3 NA 27 133 28 44 10 2 2
## 5 ansonca~ 1871 1 RC1 NA 25 120 29 39 11 3 0
## 6 armstbo~ 1871 1 FW1 NA 12 49 9 11 2 1 0
## # ... with 10 more variables: RBI <int>, SB <int>, CS <int>, BB <int>,
## # SO <int>, IBB <int>, HBP <int>, SH <int>, SF <int>, GIDP <int>
These Tables are Logically Connected
- Fields in one table “relate” to fields in one or more other tables
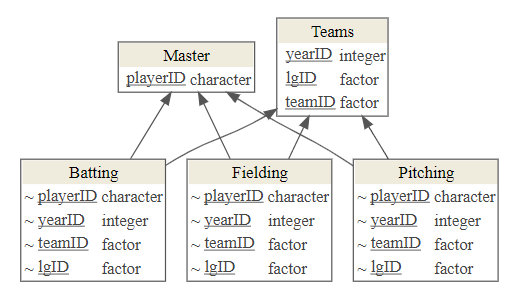
- For Lahman:
Batting,Fielding, andPitchingconnect toPeoplevia a single variable,PeopleID.Batting,Fielding, andPitchingconnect toTeamsthrough theYearID,TeamIDandlgIDvariables.
We Define Variables as “Keys” to Identify Rows and to Make Logical Connections Between Tables.
- Every Table should have a Primary key to uniquely identify (differentiate) its rows.
- Keys must be unique in their own table, i,e., only refer to one instance of an item.
- Good Data engineers create keys with no intrinsic meaning other than being a unique identifier.
- Some tables may use a combined key based on multiple columns, e.g., year, month, day together.
- The primary key from one table may appear many times in other tables.
- Example:
People$playerIDis a primary key forPeoplebecause it uniquely identifies the rows inPeople. - To check if you have identified the Primary Key fields, use
group_by(primary_key_fields)andcount()to see if there are multiple rows for any group. - If any group has more than one row, the fields are insufficient to serve as a primary key
# type is Not a Primary Key
People %>%
group_by(nameLast) %>%
count() %>%
filter(n > 1)
## # A tibble: 2,559 x 2
## # Groups: nameLast [2,559]
## nameLast n
## <chr> <int>
## 1 Aaron 2
## 2 Abad 2
## 3 Abbey 2
## 4 Abbott 9
## 5 Abercrombie 2
## 6 Abernathy 4
## 7 Abrams 2
## 8 Abreu 7
## 9 Acevedo 2
## 10 Acker 2
## # ... with 2,549 more rows
# This is a primary key
People %>%
group_by(playerID) %>%
count() %>%
filter(n > 1)
## # A tibble: 0 x 2
## # Groups: playerID [0]
## # ... with 2 variables: playerID <chr>, n <int>
- A primary key can also serve as a foreign key when present in another table.
- A Foreign key is used to identify rows in another (child) table.
- Example: In
Pitching,$playerIDis a foreign key for the other tablePeoplebecause it uniquely identifies rows inPeople. - There can be multiple rows with the same foreign key in a table, e.g.,
playerIDinPitching, soPitching$playerIDis not a primary key in Pitching
Pitching %>%
group_by(playerID) %>%
count() %>%
filter(n > 1)
## # A tibble: 6,980 x 2
## # Groups: playerID [6,980]
## playerID n
## <chr> <int>
## 1 aardsda01 9
## 2 aasedo01 13
## 3 abadfe01 10
## 4 abbeybe01 6
## 5 abbotgl01 12
## 6 abbotji01 11
## 7 abbotky01 4
## 8 abbotpa01 12
## 9 aberal01 8
## 10 abernte01 3
## # ... with 6,970 more rows
Join Functions in the dplyr Package
- Getting data from two (or more) tables requires using the primary keys and foreign keys to logically connect the tables.
- These “connections” are called joins
- The dplyr package has functions to connect or join tables so you can work with their data
- The dplyr package supports seven types of joins:
- Four types of mutating joins,
- Two types of filtering joins, and
- A nesting join.
- Many sources refer to the first table in a join function argument list as the
xtable or the left table or the “parent” table and second table in the argument as theytable or the right table or the “child” table - The primary key fields for the
y/right/child table must be able to be matched to fields in thex/left/parent table which can serve as a foreign key.
Mutating Joins: Inner, Left, Right, Full
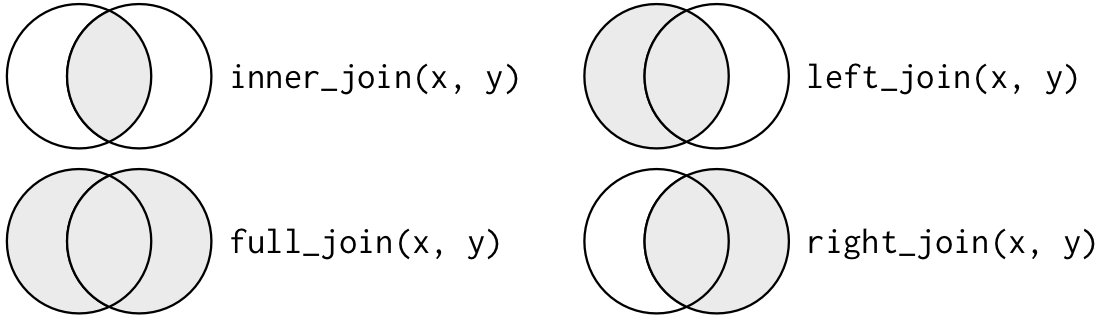
- Mutating Joins affect the rows and columns of either the
xorytable inner_join(): returns all rows from x where there are matching values in y, and all columns from x and y.
- If there are multiple matches between x and y, all combination of the matches are returned.
- Rows that do not match are not returned
left_join():returns all rows from x, and all columns from x and y.
- Rows in x with no match in y are returned but will have
NAvalues in the new columns. - If there are multiple matches between
xandy, all combinations of the matches are returned.
right_join(): returns all rows from y, and all columns from x and y.
- Rows in
ywith no match inxwill haveNAvalues in the new columns. - If there are multiple matches between
xandy, all combinations of the matches are returned.
full_join(): returns all rows and all columns from both x and y.
- Where there are not matching values, returns
NAfor the missing values.
x <- tribble(
~key, ~val_x,
1, "x1",
2, "x2",
3, "x3"
)
x
## # A tibble: 3 x 2
## key val_x
## <dbl> <chr>
## 1 1 x1
## 2 2 x2
## 3 3 x3
y <- tribble(
~key, ~val_y,
1, "y1",
2, "y2",
4, "y4"
)
y
## # A tibble: 3 x 2
## key val_y
## <dbl> <chr>
## 1 1 y1
## 2 2 y2
## 3 4 y4
inner_join(x, y)
- This is the simplest join to add
- Matches the rows of
xwith rows ofyonly when their keys are equal.
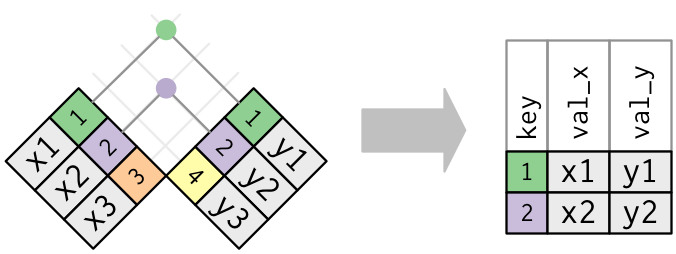
inner_join(x, y, by = "key")
## # A tibble: 2 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
- Keeps all rows that appear in both data frames.
- if the key is missing on either the left or right side, that row will be dropped from the join.
Exercise:
- Select all batting stats for players who were born in the 1980s.
People %>%
filter(between(birthYear, 1980, 1989)) %>%
nrow()
## [1] 2207
nrow(Batting)
## [1] 107429
People %>%
filter(between(birthYear, 1980, 1989)) %>%
inner_join(Batting, by = "playerID") %>%
nrow()
## [1] 12516
Outer Joins: Left, Right, Full
- Outer Joins keep all rows that appear in at least one data frame and add columns from the other table for those rows.
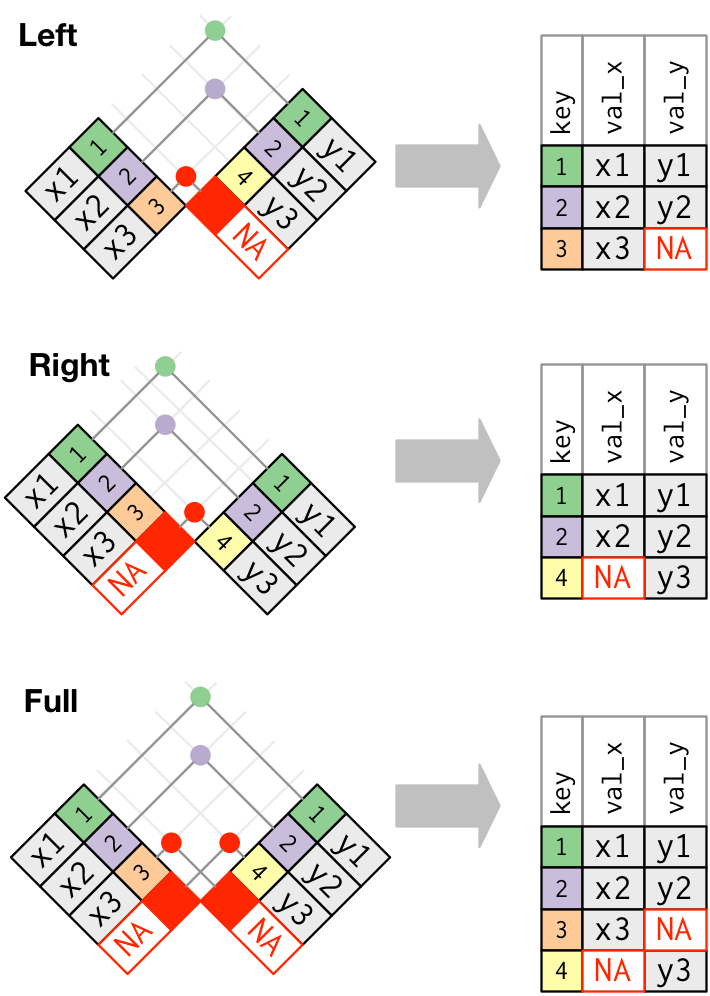
left_join(x, y)

- Keeps all rows of
xand adds columns fromy. - Puts in
NAin the newycolumns if no match. - By far the most common join
- Always use a left join unless you have a good reason not to.
left_join(x, y, by = "key")
## # A tibble: 3 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 3 x3 <NA>
right_join(x, y)

- Keeps all rows of
yand adds columns fromx. - Puts in
NAinxcolumns if no match.
right_join(x, y, by = "key")
## # A tibble: 3 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 4 <NA> y4
- Note what happens when you switch the type of join and the data frame
left_join(y, x, by = "key")
## # A tibble: 3 x 3
## key val_y val_x
## <dbl> <chr> <chr>
## 1 1 y1 x1
## 2 2 y2 x2
## 3 4 y4 <NA>
- You have flexibility to choose the order you are doing the joins and the type of joins to get the results you want from your workflow.
full_join(x, y)

- Keeps all rows of both and adds columns from
ytox. - Puts in
NAwherever there is no match - Can cause a lot of extra rows and columns with
NAsif not careful - Only use when absolutely necessary and then only after you have selected the desired variables and filtered the desired rows.
full_join(x, y, by = "key")
## # A tibble: 4 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 3 x3 <NA>
## 4 4 <NA> y4
Exercise:
- Add the team name to the
Battingdata frame.
names(Batting)
## [1] "playerID" "yearID" "stint" "teamID" "lgID" "G"
## [7] "AB" "R" "H" "X2B" "X3B" "HR"
## [13] "RBI" "SB" "CS" "BB" "SO" "IBB"
## [19] "HBP" "SH" "SF" "GIDP"
names(Teams)
## [1] "yearID" "lgID" "teamID" "franchID"
## [5] "divID" "Rank" "G" "Ghome"
## [9] "W" "L" "DivWin" "WCWin"
## [13] "LgWin" "WSWin" "R" "AB"
## [17] "H" "X2B" "X3B" "HR"
## [21] "BB" "SO" "SB" "CS"
## [25] "HBP" "SF" "RA" "ER"
## [29] "ERA" "CG" "SHO" "SV"
## [33] "IPouts" "HA" "HRA" "BBA"
## [37] "SOA" "E" "DP" "FP"
## [41] "name" "park" "attendance" "BPF"
## [45] "PPF" "teamIDBR" "teamIDlahman45" "teamIDretro"
Batting %>%
left_join(Teams, by = "teamID") %>%
select(name, everything())
## # A tibble: 8,699,563 x 69
## name playerID yearID.x stint teamID lgID.x G.x AB.x R.x H.x X2B.x
## <chr> <chr> <int> <int> <fct> <fct> <int> <int> <int> <int> <int>
## 1 Troy~ abercda~ 1871 1 TRO NA 1 4 0 0 0
## 2 Troy~ abercda~ 1871 1 TRO NA 1 4 0 0 0
## 3 Rock~ addybo01 1871 1 RC1 NA 25 118 30 32 6
## 4 Clev~ allisar~ 1871 1 CL1 NA 29 137 28 40 4
## 5 Clev~ allisar~ 1871 1 CL1 NA 29 137 28 40 4
## 6 Wash~ allisdo~ 1871 1 WS3 NA 27 133 28 44 10
## 7 Wash~ allisdo~ 1871 1 WS3 NA 27 133 28 44 10
## 8 Rock~ ansonca~ 1871 1 RC1 NA 25 120 29 39 11
## 9 Fort~ armstbo~ 1871 1 FW1 NA 12 49 9 11 2
## 10 Rock~ barkeal~ 1871 1 RC1 NA 1 4 0 1 0
## # ... with 8,699,553 more rows, and 58 more variables: X3B.x <int>, HR.x <int>,
## # RBI <int>, SB.x <int>, CS.x <int>, BB.x <int>, SO.x <int>, IBB <int>,
## # HBP.x <int>, SH <int>, SF.x <int>, GIDP <int>, yearID.y <int>,
## # lgID.y <fct>, franchID <fct>, divID <chr>, Rank <int>, G.y <int>,
## # Ghome <int>, W <int>, L <int>, DivWin <chr>, WCWin <chr>, LgWin <chr>,
## # WSWin <chr>, R.y <int>, AB.y <int>, H.y <int>, X2B.y <int>, X3B.y <int>,
## # HR.y <int>, BB.y <int>, SO.y <int>, SB.y <int>, CS.y <int>, HBP.y <int>,
## # SF.y <int>, RA <int>, ER <int>, ERA <dbl>, CG <int>, SHO <int>, SV <int>,
## # IPouts <int>, HA <int>, HRA <int>, BBA <int>, SOA <int>, E <int>, DP <int>,
## # FP <dbl>, park <chr>, attendance <int>, BPF <int>, PPF <int>,
## # teamIDBR <chr>, teamIDlahman45 <chr>, teamIDretro <chr>
- list the first name, last name, and team name for every player who played in 2018
Batting %>%
filter(yearID == 2018) %>%
left_join(People, by = "playerID") %>%
left_join(Teams, by = "teamID") %>%
select(nameFirst, nameLast, name) %>%
distinct()
## # A tibble: 2,426 x 3
## nameFirst nameLast name
## <chr> <chr> <chr>
## 1 Jose Abreu Chicago White Sox
## 2 Ronald Acuna Atlanta Braves
## 3 Willy Adames Tampa Bay Devil Rays
## 4 Willy Adames Tampa Bay Rays
## 5 Jason Adam Kansas City Royals
## 6 Austin Adams Washington Senators
## 7 Austin Adams Washington Nationals
## 8 Chance Adams New York Highlanders
## 9 Chance Adams New York Yankees
## 10 Lane Adams Atlanta Braves
## # ... with 2,416 more rows
Duplicate Keys
- One would not expect to have rows with duplicate primary keys in a table from a relational database (most enforce no duplicates on the primary key fields)
- However, data from sources without rules enforcing key uniqueness often have them.
- If you have duplicate keys the
xtable, then during a left join with aywhere there is no duplication, the rows ofyare copied multiple times into the newxdata frame.
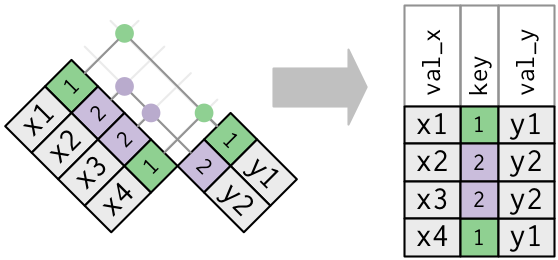
- This can be useful when you want to add additional information (rows) to
xas there can be a one-to-many relationship. - In a sense the “original”
xkey was really a foreign key with respect to the join as it did not uniquely identify the rows inxbut was a primary key fory.
x_mult <- tribble(~key, ~val_x,
##-- ------
1, "x1",
2, "x2",
2, "x3",
1, "x4")
y
## # A tibble: 3 x 2
## key val_y
## <dbl> <chr>
## 1 1 y1
## 2 2 y2
## 3 4 y4
x_mult %>%
left_join(y, by = "key")
## # A tibble: 4 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 2 x3 y2
## 4 1 x4 y1
- If you have duplicate keys in both (usually a mistake), then you get every possible combination of the values in x and y at the key values where there are duplications.
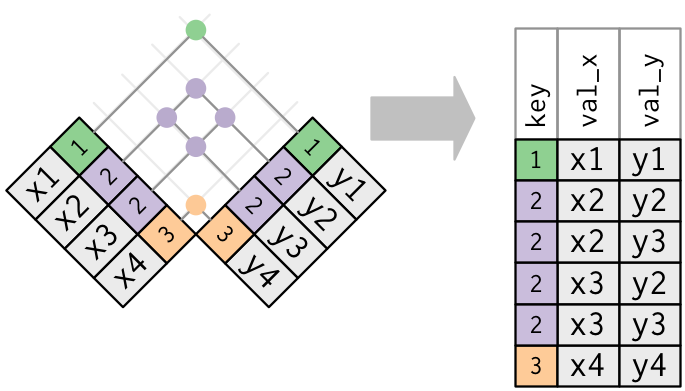
x_mult
## # A tibble: 4 x 2
## key val_x
## <dbl> <chr>
## 1 1 x1
## 2 2 x2
## 3 2 x3
## 4 1 x4
y_mult <- tribble(~key, ~val_y,
##-- ------
1, "y1",
2, "y2",
2, "y3",
1, "y4")
left_join(x_mult, y_mult, by = "key")
## # A tibble: 8 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 1 x1 y4
## 3 2 x2 y2
## 4 2 x2 y3
## 5 2 x3 y2
## 6 2 x3 y3
## 7 1 x4 y1
## 8 1 x4 y4
Filtering Joins Filter Rows in the Left (x) Data Frame:
- They do not add columns; they just filter the rows of
xbased on values iny semi_join(): returns all rows from x where there are matching key values in y, keeping just columns from x.- Filters out rows in
xthat do not match anything iny.
- Filters out rows in
- A semi-join is not the same as an inner join
- an inner join will return one row of
xfor each matching row ofy, (potentially adding multiple rows) whereas - a semi-join will never duplicate rows of
x.
- an inner join will return one row of
semi_join(x, y)

- Keeps all of the rows in
xthat have a match iny(but doesn’t add the variables ofytox).
semi_join(x, y, by = "key")
## # A tibble: 2 x 2
## key val_x
## <dbl> <chr>
## 1 1 x1
## 2 2 x2
anti_join()

anti_join(): returns all rows from x where there are not matching key values in y, keeping just columns from x.- Filters out rows in x that do match anything in y (the rows with no join).
- Drops all of the rows in
xthat do not have a match iny(but doesn’t add the variables ofytox).
In human speak, what’s in x but not y?
anti_join(x, y, by = "key")
## # A tibble: 1 x 2
## key val_x
## <dbl> <chr>
## 1 3 x3
Exercise:
- Find the 10 players with the highest number of strikeouts (
SO) from theBattingtable and assign to a new data frame. - Join the appropriate data frames to select all players from those 10 days.
ten_worst <- Batting %>%
group_by(playerID) %>%
summarize(count_strikeout = sum(SO, na.rm = TRUE)) %>%
slice_max(count_strikeout, n =10)
People %>%
semi_join(ten_worst)
## # A tibble: 10 x 26
## playerID birthYear birthMonth birthDay birthCountry birthState birthCity
## <chr> <int> <int> <int> <chr> <chr> <chr>
## 1 cansejo~ 1964 7 2 Cuba La Habana La Habana
## 2 dunnad01 1979 11 9 USA TX Houston
## 3 galaran~ 1961 6 18 Venezuela Distrito ~ Caracas
## 4 grandcu~ 1981 3 16 USA IL Blue Isl~
## 5 jacksre~ 1946 5 18 USA PA Abington
## 6 reynoma~ 1983 8 3 USA KY Pikeville
## 7 rodrial~ 1975 7 27 USA NY New York
## 8 sosasa01 1968 11 12 D.R. San Pedro~ San Pedr~
## 9 stargwi~ 1940 3 6 USA OK Earlsboro
## 10 thomeji~ 1970 8 27 USA IL Peoria
## # ... with 19 more variables: deathYear <int>, deathMonth <int>,
## # deathDay <int>, deathCountry <chr>, deathState <chr>, deathCity <chr>,
## # nameFirst <chr>, nameLast <chr>, nameGiven <chr>, weight <int>,
## # height <int>, bats <fct>, throws <fct>, debut <chr>, finalGame <chr>,
## # retroID <chr>, bbrefID <chr>, deathDate <date>, birthDate <date>
#note, you can do this same operation with filtering too:
ten_worst_vector <- ten_worst %>%
pull(playerID)
People %>%
filter(playerID %in% ten_worst_vector)
## # A tibble: 10 x 26
## playerID birthYear birthMonth birthDay birthCountry birthState birthCity
## <chr> <int> <int> <int> <chr> <chr> <chr>
## 1 cansejo~ 1964 7 2 Cuba La Habana La Habana
## 2 dunnad01 1979 11 9 USA TX Houston
## 3 galaran~ 1961 6 18 Venezuela Distrito ~ Caracas
## 4 grandcu~ 1981 3 16 USA IL Blue Isl~
## 5 jacksre~ 1946 5 18 USA PA Abington
## 6 reynoma~ 1983 8 3 USA KY Pikeville
## 7 rodrial~ 1975 7 27 USA NY New York
## 8 sosasa01 1968 11 12 D.R. San Pedro~ San Pedr~
## 9 stargwi~ 1940 3 6 USA OK Earlsboro
## 10 thomeji~ 1970 8 27 USA IL Peoria
## # ... with 19 more variables: deathYear <int>, deathMonth <int>,
## # deathDay <int>, deathCountry <chr>, deathState <chr>, deathCity <chr>,
## # nameFirst <chr>, nameLast <chr>, nameGiven <chr>, weight <int>,
## # height <int>, bats <fct>, throws <fct>, debut <chr>, finalGame <chr>,
## # retroID <chr>, bbrefID <chr>, deathDate <date>, birthDate <date>
Other Key Names
- If the primary and foreign keys names do not match, you need to specify the matching names in a vector
- Example:
left_join(x, y, by = c("a" = "b")), whereais the key inxandbis the key iny. - Here we join by single keys with different names in the
xandydata frames
library(nycflights13)
flights %>%
left_join(airports, by = c("origin" = "faa")) %>%
head()
## # A tibble: 6 x 26
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 1 517 515 2 830 819
## 2 2013 1 1 533 529 4 850 830
## 3 2013 1 1 542 540 2 923 850
## 4 2013 1 1 544 545 -1 1004 1022
## 5 2013 1 1 554 600 -6 812 837
## 6 2013 1 1 554 558 -4 740 728
## # ... with 18 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>, name <chr>, lat <dbl>,
## # lon <dbl>, alt <dbl>, tz <dbl>, dst <chr>, tzone <chr>
- If you have multiple variables acting as a combined key, specify the
byargument as a vector. - If they have the same name, you can drop the
=argument - The
,serve as an “AND” operator
Fielding %>%
left_join(Teams, by = c("yearID", "lgID", "teamID")) %>%
head()
## # A tibble: 6 x 63
## playerID yearID stint teamID lgID POS G.x GS InnOuts PO A E.x
## <chr> <int> <int> <fct> <fct> <chr> <int> <int> <int> <int> <int> <int>
## 1 abercda~ 1871 1 TRO NA SS 1 1 24 1 3 2
## 2 addybo01 1871 1 RC1 NA 2B 22 22 606 67 72 42
## 3 addybo01 1871 1 RC1 NA SS 3 3 96 8 14 7
## 4 allisar~ 1871 1 CL1 NA 2B 2 0 18 1 4 0
## 5 allisar~ 1871 1 CL1 NA OF 29 29 729 51 3 7
## 6 allisdo~ 1871 1 WS3 NA C 27 27 681 68 15 20
## # ... with 51 more variables: DP.x <int>, PB <int>, WP <int>, SB.x <int>,
## # CS.x <int>, ZR <int>, franchID <fct>, divID <chr>, Rank <int>, G.y <int>,
## # Ghome <int>, W <int>, L <int>, DivWin <chr>, WCWin <chr>, LgWin <chr>,
## # WSWin <chr>, R <int>, AB <int>, H <int>, X2B <int>, X3B <int>, HR <int>,
## # BB <int>, SO <int>, SB.y <int>, CS.y <int>, HBP <int>, SF <int>, RA <int>,
## # ER <int>, ERA <dbl>, CG <int>, SHO <int>, SV <int>, IPouts <int>, HA <int>,
## # HRA <int>, BBA <int>, SOA <int>, E.y <int>, DP.y <int>, FP <dbl>,
## # name <chr>, park <chr>, attendance <int>, BPF <int>, PPF <int>,
## # teamIDBR <chr>, teamIDlahman45 <chr>, teamIDretro <chr>
- You can specify both multiple variables and with different names if necessary
- You cannot specify one key from
xto have two different values iny- there is no “OR” operation; you have to do two joins.
Set operations
 {width = 50%}
{width = 50%}
x <- tribble(
~key, ~val,
1, "a",
1, "b",
2, "a"
)
y <- tribble(
~key, ~val,
1, "a",
2, "b",
)
Set operations are occasionally useful when you want to break a single complex filter into simpler pieces. All these operations work with a complete row, comparing the values of every variable. These expect the x and y inputs to have the same variables, and treat the observations like sets.
Union
All unique rows from x and y.

union(y, x)
## # A tibble: 4 x 2
## key val
## <dbl> <chr>
## 1 1 a
## 2 2 b
## 3 1 b
## 4 2 a
intersect
Common rows in both x and y, keeping just unique rows.

intersect(x, y)
## # A tibble: 1 x 2
## key val
## <dbl> <chr>
## 1 1 a
because table x had a dupicated row, it was removed.
setdiff
All rows from x which are not also rows in y, keeping just unique rows.

#what is in x but not in y?
setdiff(x, y)
## # A tibble: 2 x 2
## key val
## <dbl> <chr>
## 1 1 b
## 2 2 a

#what is in y but not in x?
setdiff(y, x)
## # A tibble: 1 x 2
## key val
## <dbl> <chr>
## 1 2 b
Appending Data
Sometimes you want to add rows to your data instead of adding new columns. This
happens a lot when you have new data coming in daily or weekly. To handle this
data, we will use the bind_rows() function. To see how this works, let’s look
at some play data about plant growth. In this situation, let’s say we have a
sensor sending us new readings daily. How could we combine these data into one
tibble?
march03 <- tibble::tribble(
~plant_id, ~treatment, ~height, ~temperature, ~humidity,
"k3jak", "control", 1.234156382, 82L, 0.257529979,
"df33l", "control", 1.380212809, 83L, 0.058794941,
"l122j", "treated", 1.526392762, 89L, 0.297541584,
"3kssj", "treated", 1.673373933, 80L, 0.881550707
)
march04 <- tibble::tribble(
~plant_id, ~treatment, ~height, ~temperature, ~humidity,
"k3jak", "control", 1.660054506, 92L, 0.474884172,
"df33l", "control", 1.87979144, 83L, 0.769558459,
"l122j", "treated", 1.686950799, 86L, 0.228663423,
"3kssj", "treated", 2.032178067, 87L, 0.0340041
)
march05 <- tibble::tribble(
~plant_id, ~treatment, ~height, ~temperature, ~humidity,
"k3jak", "control", 1.933533759, 89L, 0.975123013,
"df33l", "control", 1.990040605, 97L, 0.724627173,
"l122j", "treated", 1.996540851, 94L, 0.490475035,
"3kssj", "treated", 2.116540208, 93L, 0.169962639
)
march03 <- march03 %>%
mutate(date = lubridate::mdy("03/03/2021"))
march04 <- march04 %>%
mutate(date = lubridate::mdy("03/04/2021"))
march05 <- march05 %>%
mutate(date = lubridate::mdy("03/05/2021"))
plants <- march03 %>%
bind_rows(march04) %>%
bind_rows(march05)
#alternatively, we could do a "subquery" on each of these dataframes..
march03 %>%
mutate(date = lubridate::mdy("03/03/2021")) %>%
bind_rows(mutate(march04, date = lubridate::mdy("03/04/2021"))) %>%
bind_rows(mutate(march05, date = lubridate::mdy("03/05/2021")))
## # A tibble: 12 x 6
## plant_id treatment height temperature humidity date
## <chr> <chr> <dbl> <int> <dbl> <date>
## 1 k3jak control 1.23 82 0.258 2021-03-03
## 2 df33l control 1.38 83 0.0588 2021-03-03
## 3 l122j treated 1.53 89 0.298 2021-03-03
## 4 3kssj treated 1.67 80 0.882 2021-03-03
## 5 k3jak control 1.66 92 0.475 2021-03-04
## 6 df33l control 1.88 83 0.770 2021-03-04
## 7 l122j treated 1.69 86 0.229 2021-03-04
## 8 3kssj treated 2.03 87 0.0340 2021-03-04
## 9 k3jak control 1.93 89 0.975 2021-03-05
## 10 df33l control 1.99 97 0.725 2021-03-05
## 11 l122j treated 2.00 94 0.490 2021-03-05
## 12 3kssj treated 2.12 93 0.170 2021-03-05
In the example above, why do we want to bind_rows() instead of a join?
Bind rows matches column names, so if there are missing columns, they will be filled with NA in the newly added rows like the following.
a <- tibble(x = 1:3, y = c(4,5,NA))
a
## # A tibble: 3 x 2
## x y
## <int> <dbl>
## 1 1 4
## 2 2 5
## 3 3 NA
b <- tibble(y = 1:4, z = runif(4))
b
## # A tibble: 4 x 2
## y z
## <int> <dbl>
## 1 1 0.177
## 2 2 0.198
## 3 3 0.801
## 4 4 0.550
a %>%
bind_rows(b)
## # A tibble: 7 x 3
## x y z
## <int> <dbl> <dbl>
## 1 1 4 NA
## 2 2 5 NA
## 3 3 NA NA
## 4 NA 1 0.177
## 5 NA 2 0.198
## 6 NA 3 0.801
## 7 NA 4 0.550
References
- Chapter 13 of RDS.
- Data Transformation Cheatsheet
- TidyExplain GIFs
- Lahman Database visualization code
Appendix on dbplyr
While we won’t dicuss this in the lecture, this is an important reference you will likely want for working with databases in R. Feel free to run through these exercises on your own time.
- References
Motivation
- R typically works with all of the data accessible in the working memory of your computer. The more memory the larger the data set you can work with.
- In the era of big data there are many data set too large to fit into memory.
- The dbplyr package allows R to work with “remote” on-disk data stored in a database.
- This is particularly useful in two scenarios:
- Your data is already in a database on your computer or cloud drive as opposed to a .csv or rdata file.
- You have so much data it does not all fit into memory simultaneously and you need to use some external storage engine.
- If your data fits in memory, there is no advantage to putting it in a database: it will only be slower and more frustrating.
The dbplyr package
- The dbplyr functions provide a common interface to a back-end package (DBI) so you can use dplyr functions with many different databases using the same code.
- You need to install a specific back-end for the database type you want to connect to.
- Five commonly used back-ends are:
- RSQLite embeds a SQLite database.
- RMariaDB connects to MySQL and MariaDB
- RPostgres connects to Postgres and Redshift.
- odbc connects to many commercial databases via the open database connectivity protocol (ODBC).
- bigrquery connects to Google’s BigQuery.
Structured Query Language (SQL)
- SQL is a language used for many years to interact with data sets stored in relational database management systems (RDBMS).
- Common actions include: select, insert, update, or delete individual records as well as the different kinds of joins across the tables in the RDBMS
- The dbplyr package implements the most common actions people do with SQL (but SQL can do more).
- As the database back-end for dplyr, dbplyr automatically converts dplyr code into SQL
Example using RSQLite with a soccer data set
- We’ll use a soccer dataset with multiple tables to demonstrate how to use dbplyr (instead of SQL) syntax when interacting with a database.
Setup the Data and Packages
Download and unzip the database here: soccer.zip
- We’ll use the dbplyr package to interact with databases.
- dbplyr is part of the tidyverse but is not loaded as a core package with
library(tidyverse)so you have to install using the console and load it separately
install.packages("dbplyr")
library(tidyverse)
library(dbplyr)
- You’ll also need to install and load the RSQLite package.
install.packages("RSQLite")
library(RSQLite)
Two Steps to Get Started: Connect and Create Names
- Use
dbConnect()to open a connection to the database.
- Assign the output to a variable to tell R where the database is (you might need to change the path).
con <- dbConnect(drv = SQLite(), dbname = here::here("static", "data","soccer.sqlite"))
- Use
dbListTables()to list the data frames available in the connection you just created.
dbListTables(con)
## [1] "Country" "League" "Match"
## [4] "Player" "Player_Attributes" "Team"
## [7] "Team_Attributes" "sqlite_sequence"
- Use the table function,
tbl(), to create reference names (variables) for the tables incon.
- Create one for each table of interest
- Note these are not dataframes, but are lists
Team_db <- tbl(con, "Team")
Team_at_db <- tbl(con, "Team_Attributes")
Country_db <- tbl(con, "Country")
League_db <- tbl(con, "League")
Match_db <- tbl(con, "Match")
Use dplyr to Manipulate the Data
- We can now use dplyr to interact with these data frames almost like they were actually in memory (with some limitations).
head(Country_db)
head(Match_db)
Match_db %>%
select(id:away_team_goal)
names(Match_db) ## gives you the names of the list
Match_db$ops$vars
- Select the variables you want and/or the observations you want,then use
collect()to convert into a data frame in memory - Then you have access to all of the functionality of R, e.g.,
pivot_longer()andpivot_wider()(which will only work on in-memory data frames).
# Produces an error
Match_db %>%
pivot_longer(cols = contains("player"),
names_to = "Player",
values_to = "Present")
## Error in UseMethod("pivot_longer"): no applicable method for 'pivot_longer' applied to an object of class "c('tbl_SQLiteConnection', 'tbl_dbi', 'tbl_sql', 'tbl_lazy', 'tbl')"
Match <- Match_db %>%
select(id:away_team_goal) %>%
collect()
Team <- Team_db %>%
collect()
Country <- Country_db %>%
collect()
- The following will return a data frame with the home country for each team.
Match %>%
select(country_id, home_team_api_id, away_team_api_id) %>%
pivot_longer(-country_id, names_to = "home_away", values_to = "team_api_id") %>%
select(-home_away) %>%
distinct() %>%
left_join(Team, by = "team_api_id") %>% # get the team info
left_join(Country, by = c("country_id" = "id")) %>% # get Country name
select(team_long_name, team_short_name, name) %>%
rename(country_name = name) %>%
arrange(team_long_name) %>%
head()
## # A tibble: 6 x 3
## team_long_name team_short_name country_name
## <chr> <chr> <chr>
## 1 1. FC Kaiserslautern KAI Germany
## 2 1. FC Köln FCK Germany
## 3 1. FC Nürnberg NUR Germany
## 4 1. FSV Mainz 05 MAI Germany
## 5 Aberdeen ABE Scotland
## 6 AC Ajaccio AJA France
Exercise:
- Find the league_id for the
England Premier League - Extract all matches for the
England Premier Leaguefor each day in the"2010/2011"season into a new data set. - Calculate the mean of the difference between home team goals and away team goals for each day in the season.
- You’ll need to separate date and time.
- You’ll also need to use
parse_date()before you plot.
- Plot this proportion against time.
League <- League_db %>%
collect()
head(League)
## # A tibble: 6 x 3
## id country_id name
## <int> <int> <chr>
## 1 1 1 Belgium Jupiler League
## 2 1729 1729 England Premier League
## 3 4769 4769 France Ligue 1
## 4 7809 7809 Germany 1. Bundesliga
## 5 10257 10257 Italy Serie A
## 6 13274 13274 Netherlands Eredivisie
League %>%
filter(name =="England Premier League")
## # A tibble: 1 x 3
## id country_id name
## <int> <int> <chr>
## 1 1729 1729 England Premier League
subMatch <- Match_db %>%
select(league_id, season, date, home_team_goal, away_team_goal) %>%
filter(league_id == 1729, season == "2010/2011") %>%
collect()
ave_diff <- subMatch %>%
separate(col = "date", into = c("date", "time"), sep = " ") %>%
select(-time) %>%
group_by(date) %>%
summarize(mean_diff = mean(home_team_goal - away_team_goal)) %>%
mutate(date = parse_date(x = date, format = "%Y-%m-%d"))
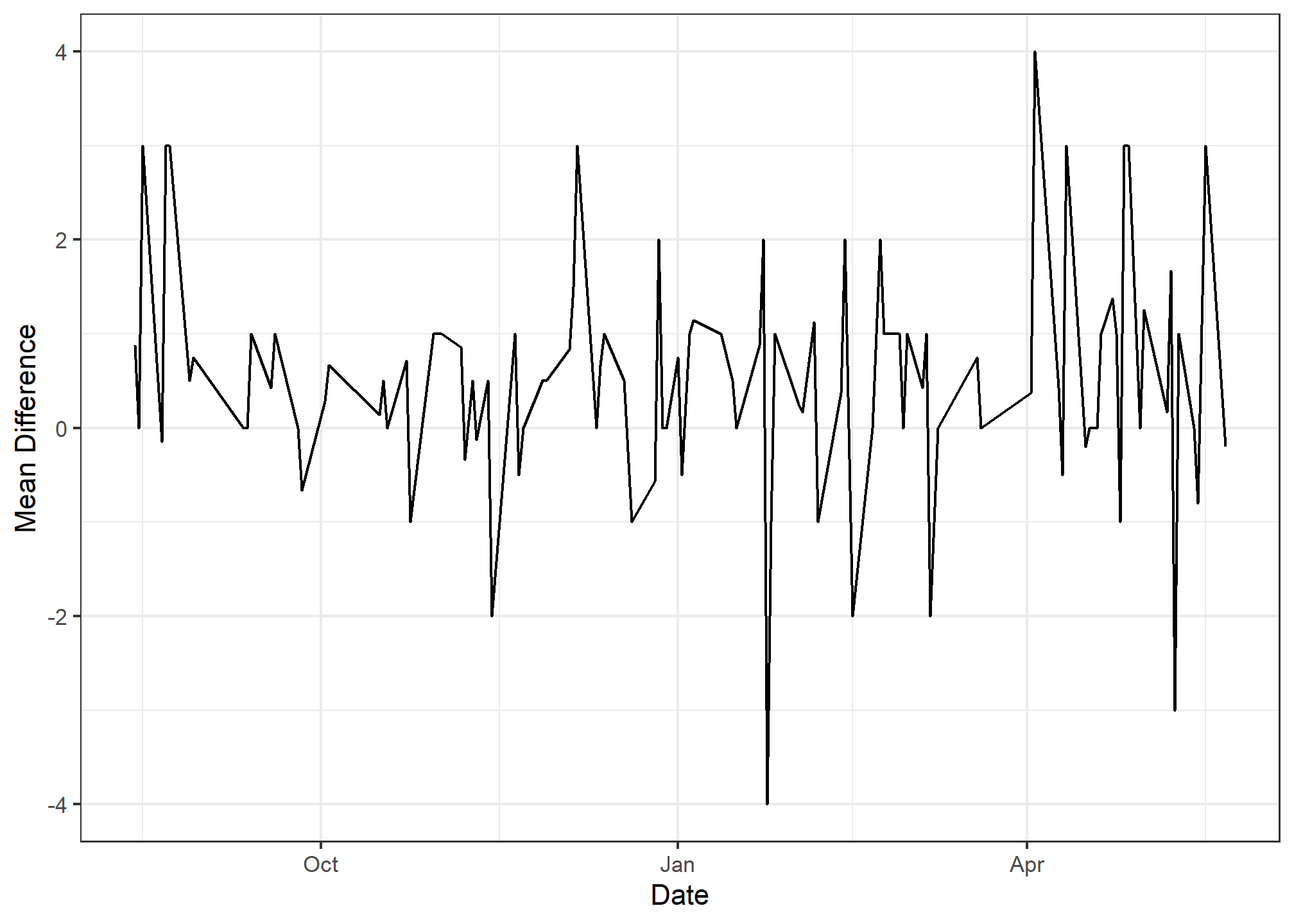
- Your plot should look like this:
ggplot(data = ave_diff, mapping = aes(x = date, y = mean_diff)) +
geom_line() +
xlab("Date") +
ylab("Mean Difference") +
theme_bw()
You can also visualize your database relationship with the dm package. While the primary and foreign keys are generally recognized, in our case we need to add them manually. You can find more information about the dm package here
install.packages("dm")
library(dm)
dm <- dm_from_src(con, learn_keys = FALSE) %>%
dm_add_pk(Country, id) %>%
dm_add_pk(League, id) %>%
dm_add_pk(Team, team_api_id) %>%
dm_add_pk(Team_Attributes, id) %>%
dm_add_pk(Match, id) %>%
dm_add_pk(Player, id) %>%
dm_add_pk(Player_Attributes, id) %>%
dm_add_fk(League, country_id, Country) %>%
dm_add_fk(Match, country_id, Country) %>%
dm_add_fk(Match, home_team_api_id, Team) %>%
dm_add_fk(Match, away_team_api_id, Team) %>%
dm_add_fk(Match, league_id, League) %>%
dm_add_fk(Player_Attributes, id, Player) %>%
dm_add_fk(Team_Attributes, team_fifa_api_id, Team) %>%
dm_add_fk(Team_Attributes, team_api_id, Team)
dm %>%
dm_draw()